Glycan3DB
A microbial carbohydrate structure database
Introduction
Molecular database systems are incredibly important in supporting the research into microbial carbohydrates. However, many of the currently available options in glycomics are littered with issues in both their functionality, and their usability. Microbial carbohydrates are so important to research as they are present on the shell of many disease-causing viruses and bacteria. They also play a role in cancer metastasis. Understanding and researching them has potential impacts in drug and vaccine research at both an academia and an industry level. Having a store of information on their structure is a functional and useful way of speeding up research, through linking to more in depth scientific papers, as well as easy access to information and fast classification. This database focuses on Klebsiella pneumoniae data but is open to expansion for any microbial carbohydrate structure though the suggestion form.
Glycan3DB structure: Main Page
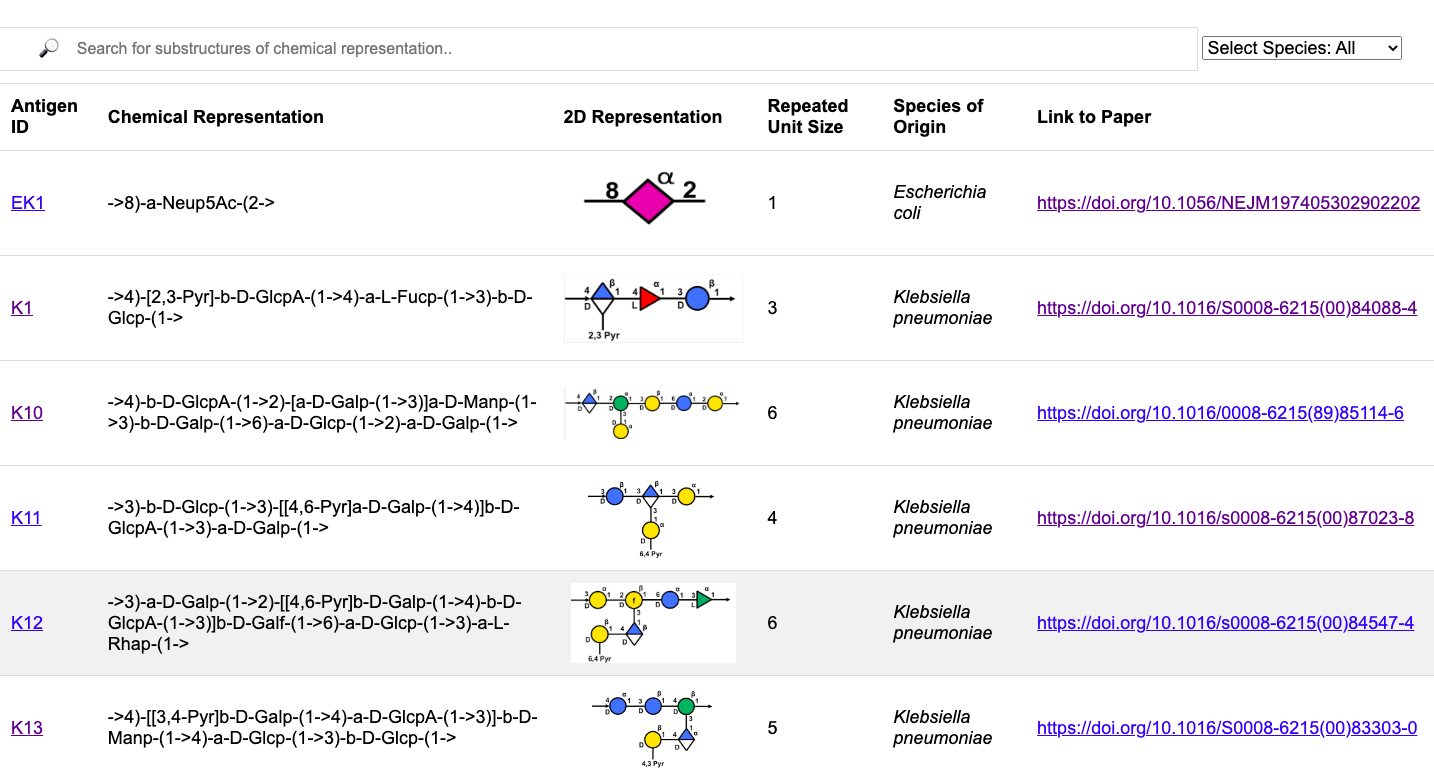
Glycan3DB has two main elements. A main "table" page and a detailed infocard page on each carbohydrate in the database. A user enters onto the main page and from there can search carbohydrates by their CASPER string format, using one or multiple substructures within the carbohydrate. The data they are shown is the antigen ID, the CASPER string format, the 2D Symbol Nomenclature for Glycans format, the repeated unit size, the species of origin and a link to the paper where the antigen is further researched.
The main page is shown below.
Glycan3DB structure: Infocard and 3D visualisation
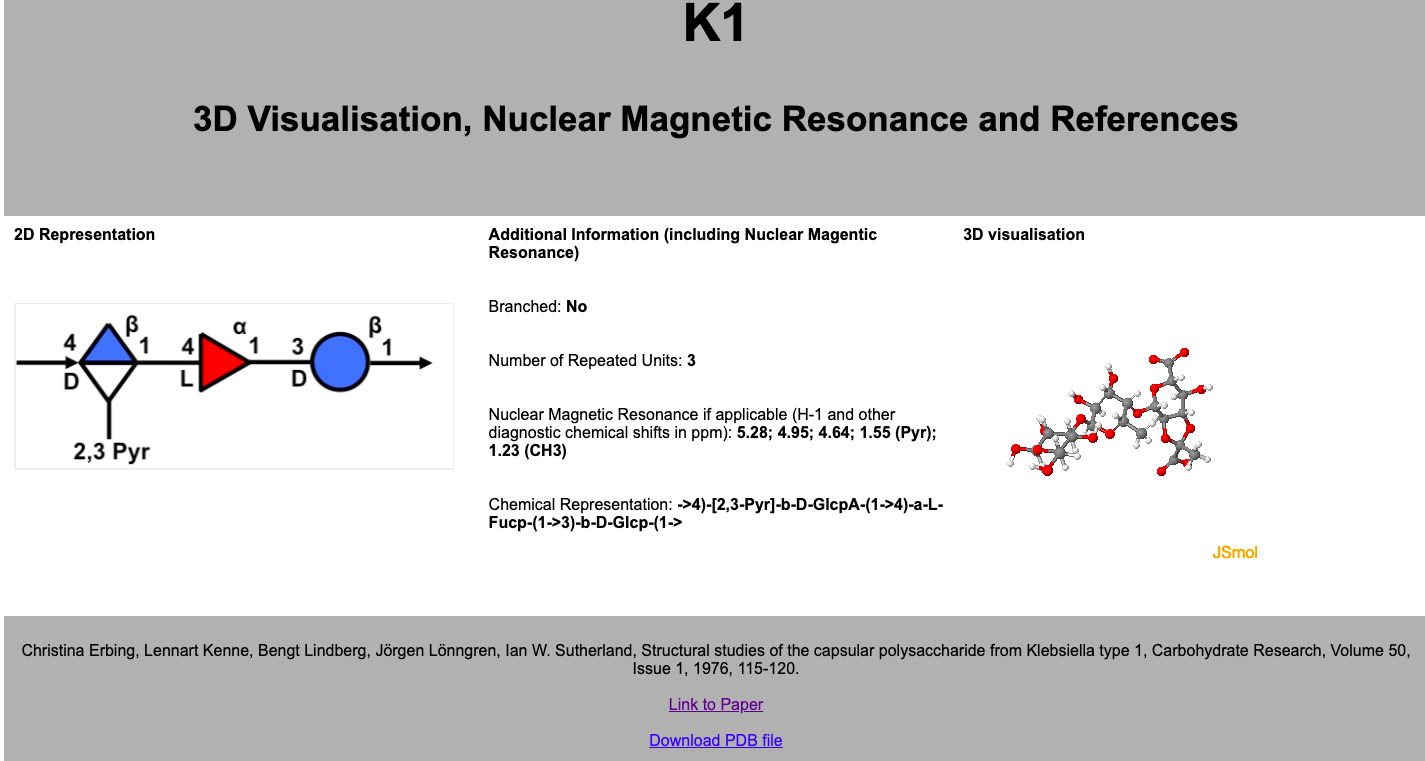
The infocard is shown below. This infocard contains more in depth data on the antigen including a 3D "ball and stick" representation which is embedded using JSmol, a 3D molecule viewer Javascript package. In also contains the references of the paper which disusses the antigen and Nuclear Magnetic Resonance data if it exists for that antigen. The 2D SNFG image of the antigen is also larger and more usable in this page.
Non-functional requirements
It was imporant for the success of this project to define what requirements we had before the project start. A few key factors were identified. This icluded follow some design rules from Ben Schneiderman. The three rules we followed were Overview, Zoom and Filter, and Details on Demand. Overview was seen as extremely important. In alternative databases, on arrival to the database, it is hard to know exactly what data is hosted in the database and thus we created an information carousel which shows what data is hosted it Glycan3DB. This is hosted above the table on the main page and is shown below

User and Expert Testing
After some iterations of development, students and experts in the field of carbohydrates were asked to test the functionality and usability of Glycan3DB. These tests were fruitful in changing the look nd structure of the webpage, as well as adding some new features, such as the ability to search by multiple substructures, and the incorporation of a help page. They also gave notes on the ability to download the 3D coordinates file, which was added to the infocard
Extensibility
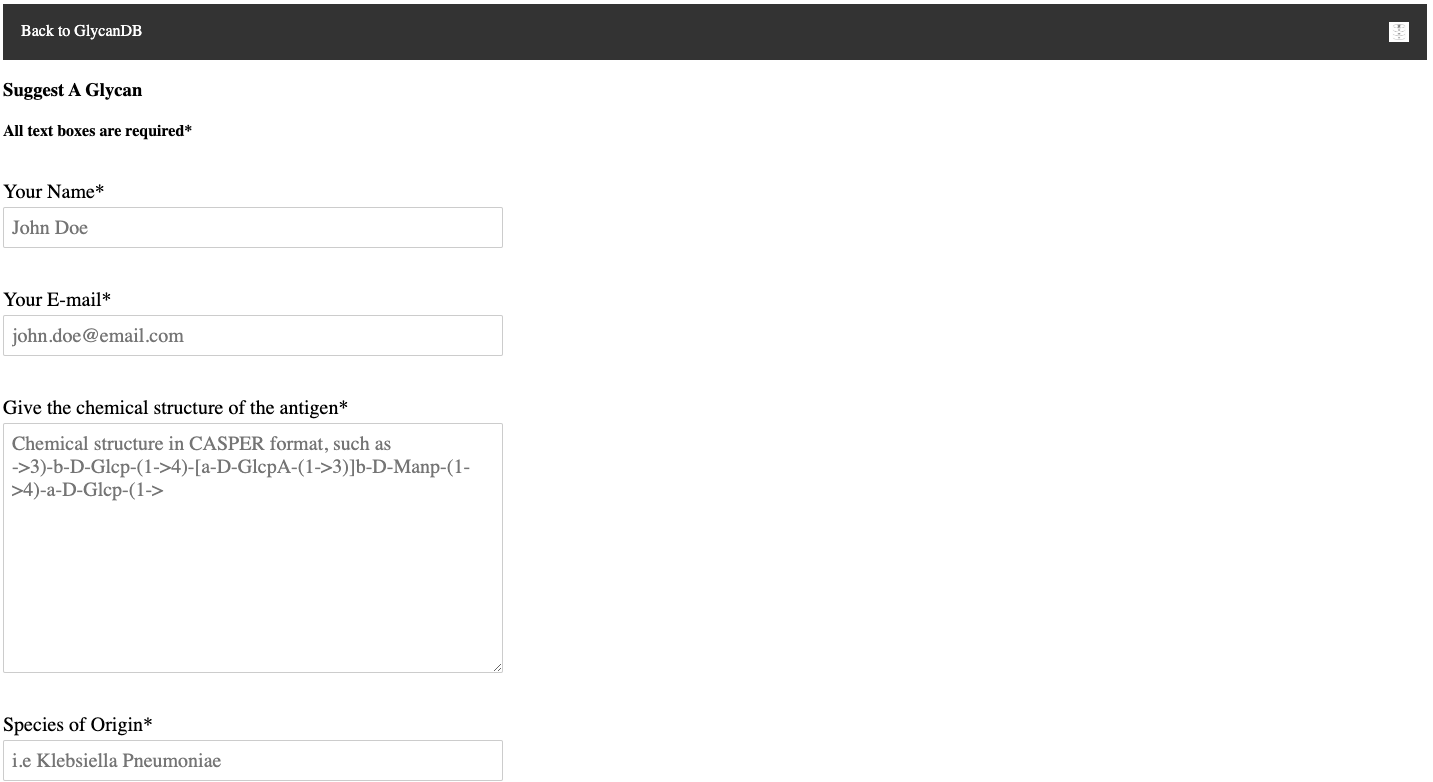
It was important for the success of Glycan3DB to be extensible by the community so a suggestion form was implemented. This form allows users of the webpage to suggest new antigens, with the suggestion being emailed to the site owner, through the use of the php mail function.
Conclusions
The main aims of development and implementation of a general, usable and extensible microbial structures database dashboard were acheived. Through many iterations, additions and tests by myself, experts and potential users, Glycan3DB was developed with all functional goals acheived. It is a system with potential for further development in the aesthetic area, as well as incorporation of new data and new species. It will give researchers and vaccine developers a platform for easy research and classification of antigens for Klebisella pneumoniae, and will only become more useful with further additions of species and antigens. This can be found at https://glycan3db.cs.uct.ac.za/.